By taking advantage of the natural tendencies of the S. pyogenes CRISPR-Cas9 system (clustered, regularly interspaced, short palindromic repeats–CRISPR-associated protein 9), the Cas9 enzyme can be used to perform site‑specific genome editing in eukaryotic cells. The WT Cas9 protein derived from S. pyogenes contains two endonuclease domains (RuvC and HNH) that function together to generate blunt-ended double stranded breaks (DSBs) by cutting opposite strands of double-stranded DNA (dsDNA). While in general, DSBs are beneficial for genome editing, it has been found that creating double stranded breaks with overhangs using Cas9 nickase enzymes greatly reduces off-target effects [1]. By taking advantage of this feature, if the correct parameters are in place, one can get both increased editing efficiency and decreased off-target effects.
How are breaks in DNA repaired?
There are two main pathways that are responsible for repairing breaks in DNA. The non-homologous end joining (NHEJ) pathway and high fidelity homology-directed (HDR) repair pathway. For situations in which minimal off-target activity and efficient, high fidelity genome editing is important, then approaches that promote HDR repair are favored over those that result in repair via NHEJ. NHEJ is much more likely to result in errors and is much less efficient while HDR allows for a more precise editing event using a donor template [2].
What is a nickase?
The WT Cas9 protein contains two endonuclease domains (RuvC and HNH) that work together in generating blunt-ended DSBs. In S. pyogenes, Cas9 creates this DSB 3 bp upstream of the protospacer-adjacent motif (PAM) sequence where the HNH domain cleaves the complementary DNA strand and the RuvC domain cleaves the non-complementary domain simultaneously [3]. To create a single nick in only one strand of the target dsDNA, one of these two endonuclease domains is inactivated, which in turn generates the Cas9 nickase mutant. The RuvC mutant (Cas9 D10A) generates a nick on only the targeting strand (gRNA complementary), while the HNH mutant (Cas9 H840A) generates a nick on only the non-targeting strand (gRNA non-complementary) (Figure 1).
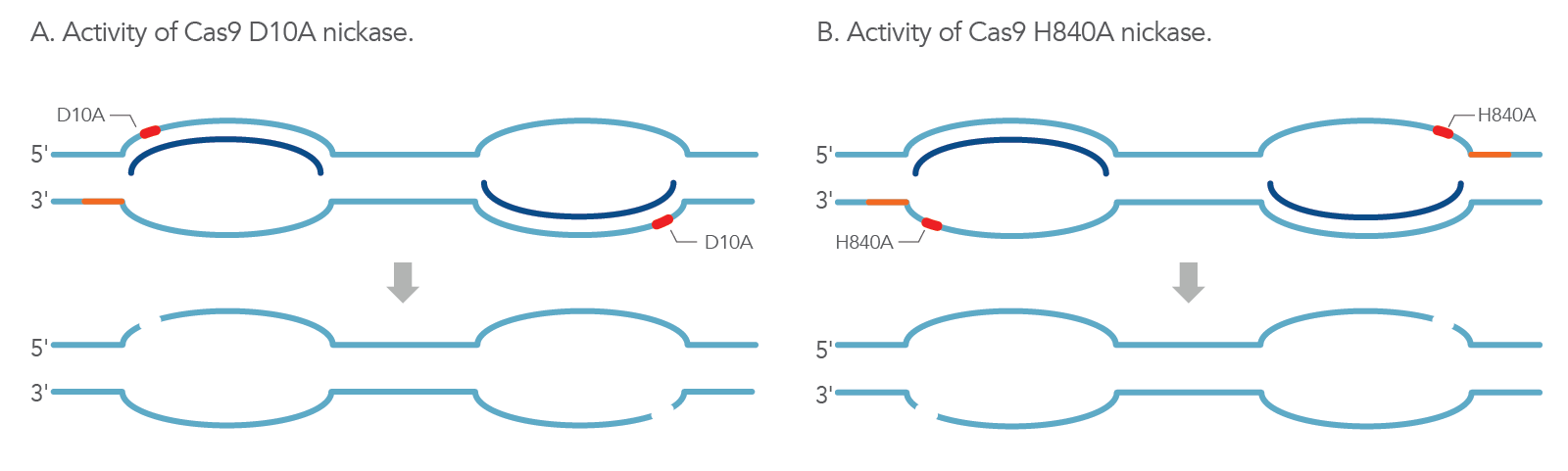
How can nickases be used for HDR?
To generate DSBs with overhangs usings Cas9 nickases, a pair of gRNAs targeting opposite DNA strands is used. The NGG protospacer adjacent motif (PAM) of the two guides can either face outwards in what is called a PAM-out orientation or towards each other in the PAM-in direction (see Figure 2), but it has been shown that with both Cas9 D10A and Cas9 H840A, the guide pairs in a PAM-out configuration result in the highest editing efficiency. In addition, Cas9 D10A is generally more potent at mediating HDR events compared to Cas9 H840A. Simultaneous nicking using a pair of gRNAs extends the number of specifically recognized bases in the target site allowing for a reduction in off-target effects [1]. Couple this with a PAM-out orientation using cleavage sites that are optimal distance apart (40–70 bp for Cas9 D10A and 50–70 bp for Cas9 H840A) using RNPs formed in separate reactions (as opposed to mixing them all in a single tube) and nickases can successfully be used for HDR mediated CRISPR editing. Read more about nickases and how to use them for editing here.
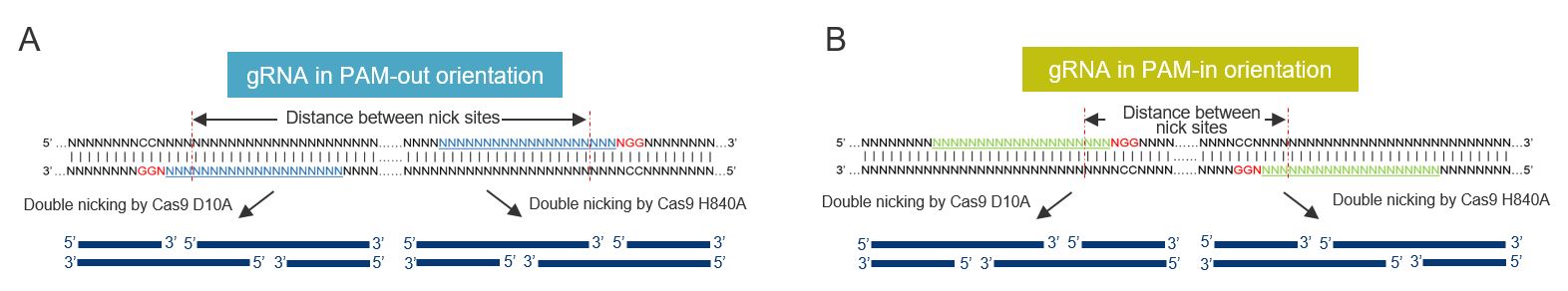
Additional ways to improve Cas9 nickase HDR
HDR efficiency can be improved by using donor oligos keeping in mind donor template design parameters, such as length of the homology arms, symmetry around insertion site, strand preference, and choice of nickase variant (D10A vs. H840A).
Cas9 D10A results in better HDR than Cas9 H840A, despite having total editing efficiencies that are overall comparable [4,5]. When creating small insertions with single-stranded oligos using Alt-R™ HDR Donor Oligos as well as the Alt-R HDR Enhancer V2 , homology arm lengths of 30–60 bases result in increased HDR rates (see Application Note). Note that there is no advantage when the ssODN template was designed asymmetrically versus symmetrically and ssODN donor templates can be designed using the sequence of either strand of the template DNA. In fact, designing and testing templates complementary to both strands is recommended whenever possible, as strand preference differs between designs and is not always predictable.
For large insertions (>120 bp), using dsDNA repair templates are advised. Alt-R™ HDR Donor blocks allow for larger knock-ins while still decreasing the off-target risk. By designing these donors with homology arms between 200–300 bp, and using them in combination with the Alt-R HDR Enhancer V2, efficient knock-in and improved HDR efficiency of sequences of up to 2000 bp can be achieved.
Design rules for consideration
- Create staggered DSBs generate overhangs to minimize off-target effects by using nickases and paired gRNAs.
- Identify paired gRNAs close to the desired sequence change with the recommended PAM-out orientation and distance between cleavage sites based upon selected nickase (40–70 bp for D10A and 50–70 bp for H840A).
- Design the gRNAs in a PAM-out orientation to achieve higher editing efficiency. The HDR Design Tool can be used to design these sequences.
- Use of donors increases efficiencies— select the type based upon the size of insert and use the HDR Design Tool to
help design these sequences.
- ssODN such Alt-R™ HDR Donor Oligos for small inserts (less than 120 bp)
- dsDNA repair templates for larger inserts such as Alt-R™ HDR Donor blocks (>120 bp)
- Cas9 D10A is more potent at mediating HDR than Cas9 H840A.
- Use of the Alt-R HDR Enhancer V2 increases HDR efficiency when used along with a donor template.
Additional notes to keep in mind
As best practice, the activity of each crRNA in a pair should be verified independently with WT Cas9 before being used in experiments with Cas9 nickases. Test the editing efficiency of the gRNA pairs and choose the pair with the highest editing efficiency for best results. Activity is highest when RNP complexes are formed separately for each gRNA before co-delivery, rather than RNP formation as a single reaction containing both paired gRNAs. Finally, these design recommendations have been incorporated in the HDR Design Tool which greatly simplifies the guide RNA and donor template design process.

