Custom Guide RNA Synthesis for CRISPR Applications
Guide RNAs for novel Cas enzymes (e.g., prime editing or the Cas13 family)
Our well-established expertise in oligo synthesis combined with an active CRISPR research and development program are what make us a trusted vendor for custom gRNA synthesis.
Ordering
CRISPR Custom Guide RNAs
We offer chemically synthesized and modified custom guide RNAs for common as well as specialized research applications. Alt-R Custom Guide RNAs are ideal for prime editing (pegRNA) projects, CRISPR-Cas13 applications, and most alternative CRISPR-Cas systems.
- Fast shipping (usually 3–6 business days)
- A range of modification and purity options
- Multi-scale delivery in tubes or plates
- gRNA available up to 175nt in length
We also offer a complete selection of Cas9 guide RNAs (sgRNA, crRNA, and tracrRNA) as well as Cas12a guide RNAs.
All our CRISPR guide RNAs are available with chemical modifications that increase stability, potency, and/or nuclease resistance. Some modifications can decrease unwanted immune responses in your experiments. Guide RNAs in tubes are available in either HPLC-purified or standard desalt formats. Pricing table reflects standard purification. HPLC pricing will be shown in cart when selected.
Standard desalt purification
HPLC purification
HPLC-purified sgRNAs are ideal for translational applications that require higher specificity and efficiency, such as gene editing in primary cells and in vivo models. HPLC gRNAs ship in as little as 12 business days. Actual TAT will vary by guide length and modification pattern. Your gRNA purity is only as accurate as your QC method. IDT has one of the most stringent QC methods on the market so you can be confident in the purity of your gRNAs every time.
IDT's purified guide RNA manufacturing process has been optimized for the unique challenges of synthesizing these molecules. By increasing manufacturing reliability at all steps of the process, adapting manufacturing protocols for long RNA oligonucleotides, and dedicating more specialized resources to these products, IDT can deliver higher purity guide RNAs faster than ever before.
|
Available custom guide RNA modifications* (up to 175nt) |
|
|---|---|
|
Available in any combination |
RNA, 2'O-Methyl, phosphorothioate bonds |
|
One modification type per guide |
DNA, 2' Fluoro, Affinity Plus Locked Nucleic Acids |
*some modifications will incur an additional charge displayed at checkout
Don’t see what you’re looking for?
Have questions for our CRISPR experts? Your time is valuable and we’ll prioritize your inquiry. Click on “Request a consultation” to provide brief information about your project, and we’ll be in touch to discuss it ASAP.
Request a consultationProduct details
Flexibility to meet a wide range of guide RNA needs
Alt -R CRISPR Custom Guide RNAs are chemically synthesized and modified. These guide RNAs include pegRNA for the prime editing system as well as guide RNAs for the Cas13 family of enzymes and other unusual Cas systems.
Prime editing guide RNA (pegRNA)
The laboratory of David Liu (Broad Institute of MIT and Harvard, Cambridge, MA) has published a CRISPR genome editing technique called prime editing [1]. This utilizes a Cas9 H840A nickase fused to a reverse transcriptase, in combination with a long guide RNA (prime editing guide RNA or pegRNA). The pegRNA encodes the new sequence and allows transcription to introduce the desired mutations. This system may have great promise for research geared towards the development of new therapies for genetic diseases. It is an exciting complement to existing CRISPR editing systems and may even be an improvement in many cases.
Cas13 guide RNA
The CRISPR-Cas13 family of enzymes targets and cuts RNA instead of DNA. Papers from the Feng Zhang lab at the Broad Institute and the Stanley Qi lab at Stanford University demonstrate the potential of CRISPR-Cas13 for SARS-CoV-2 identification as well as the research studies geared towards the development of COVID-19 therapies [2,3].
Product data
HPLC purified sgRNA
IDT's purified guide RNA manufacturing process has been optimized for the unique challenges of manufacturing these molecules. By increasing manufacturing reliability at all steps of the process, adapting manufacturing protocols for long RNA oligonucleotides, and dedicating more specialized resources to these products, IDT can deliver higher purity guide RNAs faster than ever before.
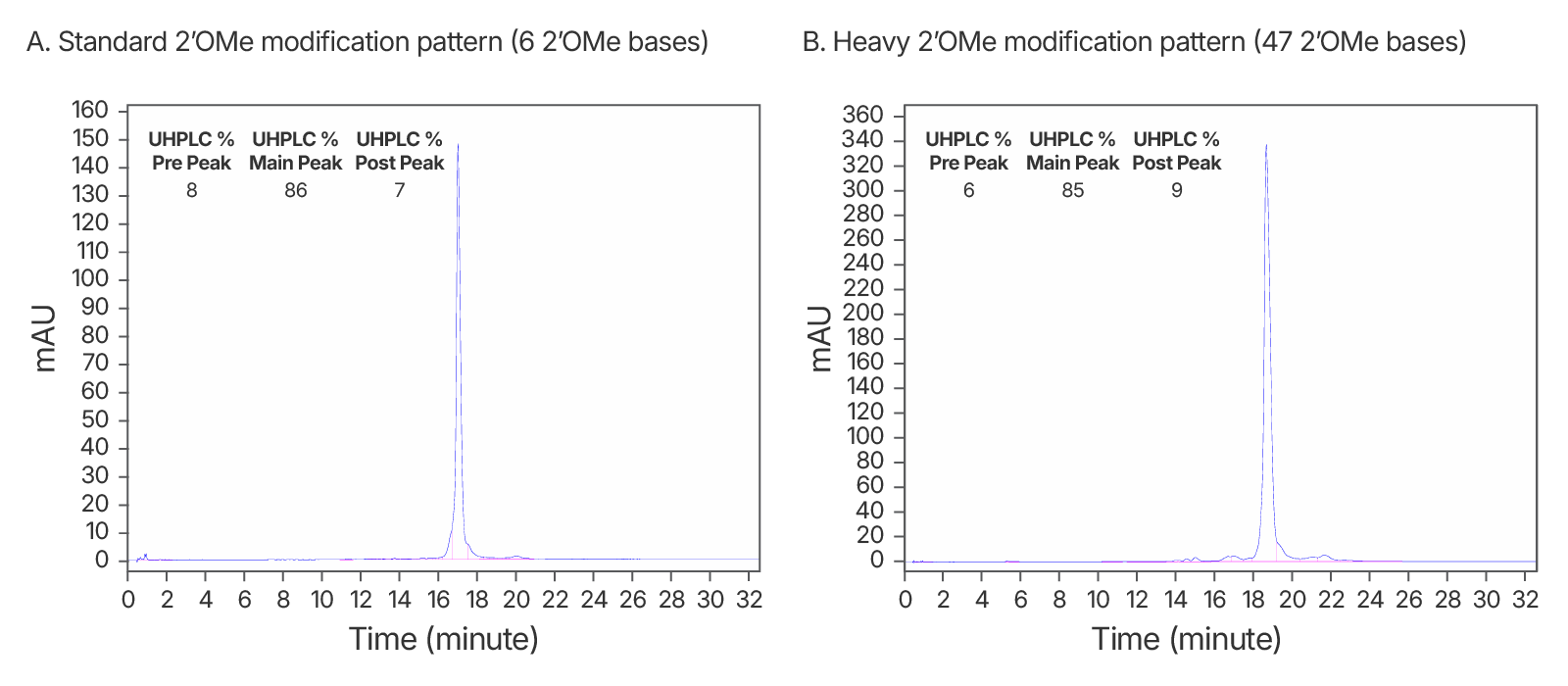
Figure 1. High quality IDT HPLC-purified gRNAs with standard modification and heavy 2’OMe modification patterns. Consistently >80% purity via UHPLC with both modification patterns. (A) An IDT chromatogram of an HPLC-purified 100 nt gRNA with a standard modification pattern (6 2'OMe, 6 Phosphorothioate Bonds). The estimated purity is 86%. (B) An IDT chromatogram of an HPLC-purified 100 nt gRNA with a heavy modification pattern (47 2'OMe, 6 PS bonds). The estimated purity is 85%.
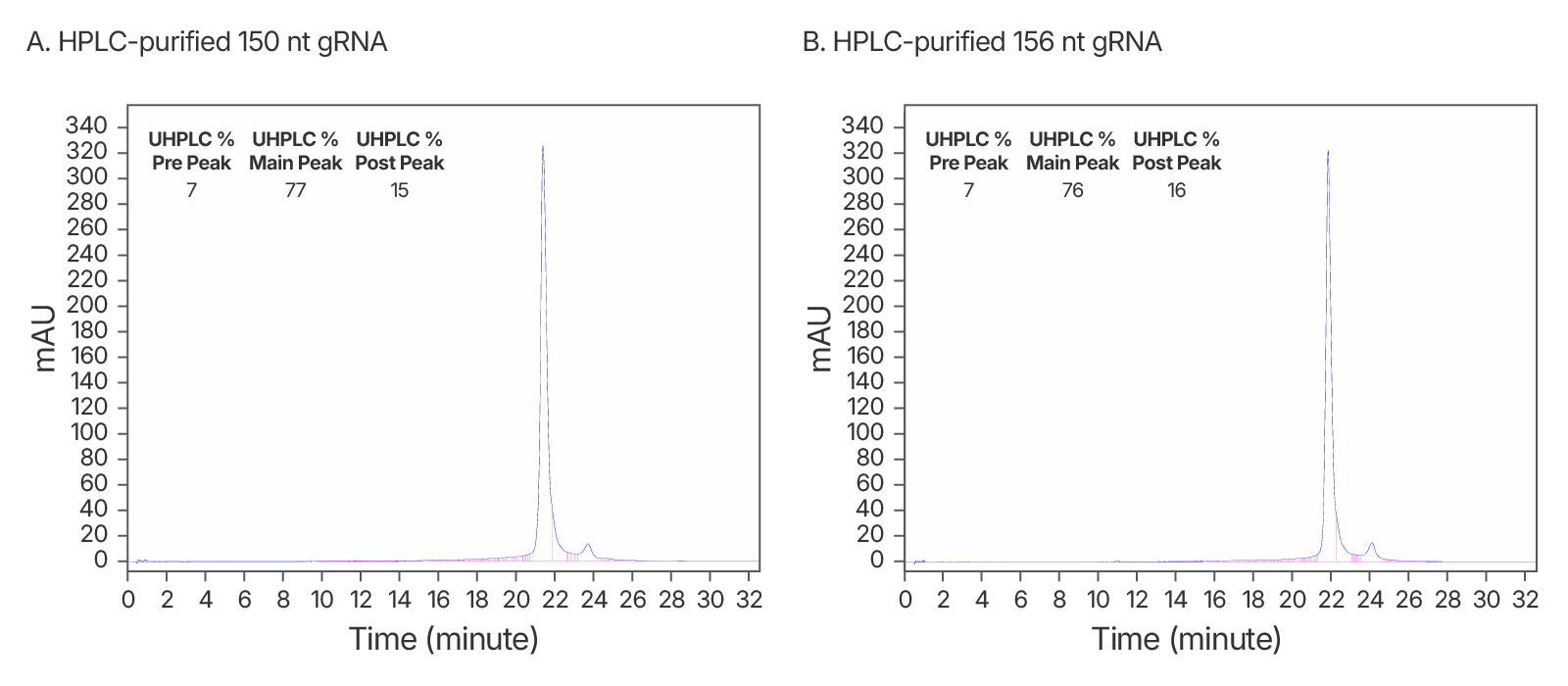
Figure 2. High quality IDT HPLC-purified long gRNAs (A) High purity maintained at >150 nt. An IDT chromatogram of an HPLC-purified 150 nt gRNA with a standard modification pattern (6 2'OMe, 6 Phosphorothioate Bonds). The estimated purity is 77%. (B) An IDT chromatogram of an HPLC-purified 156 nt gRNA with a standard modification pattern (6 2'OMe, 6 Phosphorothioate Bonds). The estimated purity is 76%.
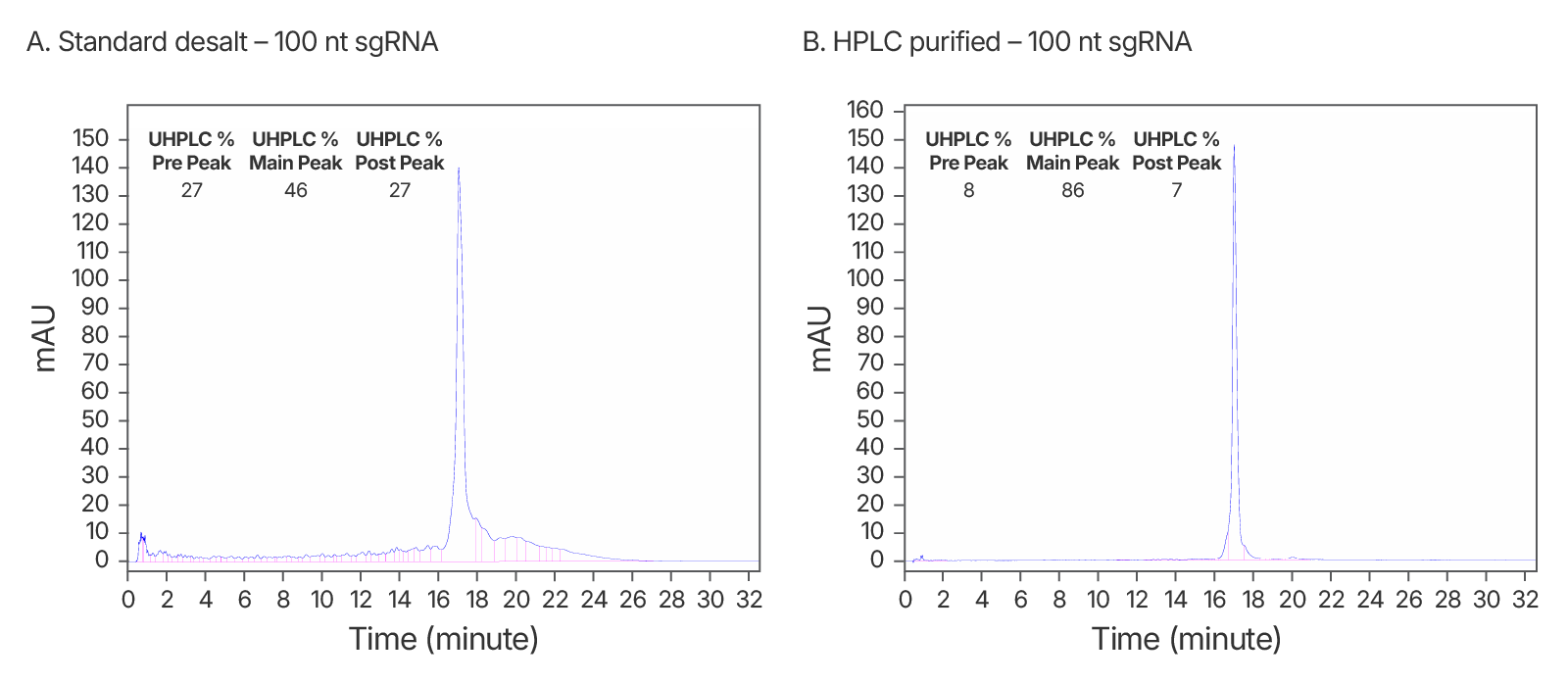
Figure 3. Purity analysis of IDT standard desalt vs. HPLC-purified sgRNAs. (A) ~2-fold increase in purity observed after HPLC purification. An IDT chromatogram of a standard desalt 100 nt gRNA with a standard modification pattern (6 2'OMe, 6 Phosphorothioate Bonds). The estimated purity is 46%. (B) An IDT chromatogram of an HPLC-purified 100 nt gRNA with a standard modification pattern (6 2'OMe, 6 Phosphorothioate Bonds). The estimated purity is 86%.
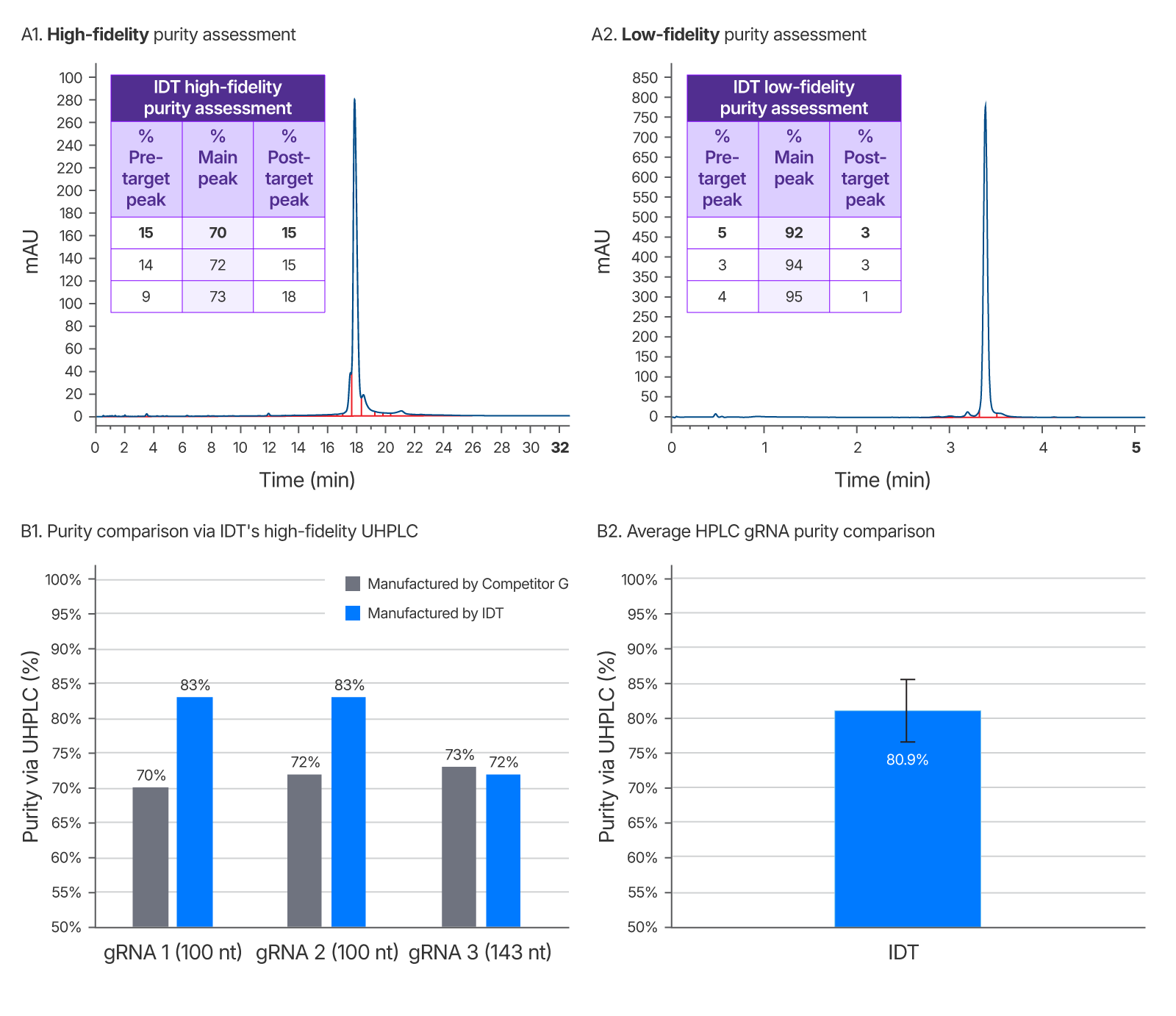
Figure 4. IDT’s gRNA UHPLC purity assessment method vs a leading competitor. Left chromatogram (A1) represents purity of competitor G sgRNA using IDT’s high-fidelity QC method. Right chromatogram (A2) is purity as reported by a low-fidelity QC method performed at IDT which does not allow enough time for sample separation and indicates artificially high purity. This low-fidelity method was developed to demonstrate the importance of a longer run time for purity accuracy. It was repeated for 3 guides ranging from 100–143 nt (one representative chromatogram shown). (B1) Average gRNA purity of three gRNAs of identical sequence from a competitor and IDT using IDT’s high-fidelity QC to eliminate inflated purity estimates. (B2) Average purity of IDT HPLC gRNAs with a range of chemical modification patterns n = 102.
Resources
Frequently asked questions
Do you use the same purification methods for RUO and CGMP guide RNA?
While some aspects of the purification process are similar between RUO and CGMP grade guide RNA, methods may not be identical. If you have specific requirements about the purification method used for your order, please contact our CGMP manufacturing team.
Can I order HPLC purified guide RNAs in Engineering Run or CGMP grades?
Yes. Please visit our CGMP gRNA web page for more information. It’s easy to speak to our experts for a CGMP consultation.
Do you perform ligation or another method to make high purity pegRNAs?
IDT does not perform any ligation reactions or click chemistry to provide long guide RNAs. However, we offer the components for these methods. If you have any questions about how to design the components for ligation in your lab, please contact us.
How can I order HPLC purified guide RNAs with different lengths, modifications, and scales?
Certain yields and combinations of modifications are available for ordering on the CRISPR-Cas9 and Custom guide RNA ordering pages. If you don't see yield you are looking for, please contact us.
When should I use an HPLC-purified guide RNA instead of standard desalt?
Standard dealt purification is suitable for routine laboratory experiments, such as initial screening or proof-of-concept studies, where ultra-high purity is not as critical. We recommend you consider HPLC purification for applications where high specificity and efficiency are critical, such as in vivo experiments or translational applications.
What CRISPR nucleases are available from IDT?
We are rapidly expanding our offerings—please check back regularly for additional options.
What purifications are supported through the Alt-R™ CRISPR Custom Guide RNA ordering tool?
Should pegRNAs for CRISPR prime editing include chemical modifications?
How can users include modifications or base types other than what is available in the Alt‑R™ CRISPR Custom Guide RNA ordering tool?
For modifications or base types other than what is available in the Alt-R™ CRISPR Custom Guide RNA ordering tool, please contact us for additional information and recommendations.
What bases and chemical modifications are supported through the Alt-R™ CRISPR Custom Guide RNA ordering tool?
References
- Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. Dec 2019;576(7785):149-157.
- Lin X, Liu Y, Chemparathy A, Pande T, La Russa M, Qi LS. A comprehensive analysis and resource to use CRISPR-Cas13 for broad-spectrum targeting of RNA viruses. Cell Rep Med. 2021;2(4):100245.
- Patchsung M, Jantarug K, Pattama A, et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed Eng. 2020;4(12):1140-1149.