xGen™ Metagenomics Amplicon Panels
Research identities in mixed microbial community
16S v2 and ITS1 panels facilitate NGS research on complex microbial communities using a single primer pool targeting the 16S rRNA gene or ITS1 region. These panels can be customized with additional targets including antibiotic resistance or virulence genes.
xGen NGS—made for metagenomics.
Ordering
- The xGen 16S rRNA v2 Amplicon Panel targets V1–V9 variable regions of the 16S rRNA gene
- The xGen ITS1 Amplicon Panel targets the internal transcribed spacer regions of the rRNA genes
- Integrated library normalization enables streamlined library balancing and pooling without the need to quantify samples
- Unique amplicon chemistry generates diverse clusters without PhiX or phased primers, leading to more efficient sequencing (Figure 4)
- Compatible with Illumina® sequencers and read lengths
- Simple, single tube, 2-hour workflow
Transform Your NGS Workflow with Automation
Looking to streamline your NGS workflows? Discover how automation can enhance efficiency and consistency in your lab with our NGS Automation solutions.
Request a consultation
Want to learn more about our xGen™ Amplicon Panels—developed with super amplicon technology—and how to customize your panel or spike-in genes in our predesigned panels? Your time is valuable—we’ll prioritize your inquiry and be in touch to discuss it ASAP.
Request a consultationProduct details
The xGen 16S Amplicon Panel v2 and the xGen ITS1 Amplicon Panel offer a reliable NGS workflow that provides coverage and NGS quality data on Illumina® sequencing platforms. These kits leverage multiplex PCR technology, enabling library construction from DNA using tiled primer pairs to target V1-V9 variable rRNA regions and the ITS1 region, each with a single pool of multiplexed primer pairs.
These xGen Amplicon Panel kits utilize multiple overlapping amplicons in a single tube, using a rapid, 2-hour sequencing workflow to prepare ready-to-sequence libraries for research studies (Figure 1). The single pool multiplex PCR workflow generates robust libraries, even from low-input quantities. The libraries may be quantified with conventional methods such as Qubit® (Thermo Fisher Scientific) or Bioanalyzer® (Agilent) and normalized by manual pooling or may be normalized enzymatically with the included xGen Normalase™ reagents. (Note: using the Normalase reagent adds additional time to complete the workflow).
In addition, both the xGen 16S Amplicon Panel v2 and the xGen ITS1 Amplicon Panel facilitate NGS analysis of complex microbial communities (e.g., bacteria, archaea, fungi) using single primer pools that target the 16S rRNA gene (variable regions 1–9) and ITS1 region, respectively. The panels can be customized with additional targets, including antibiotic resistance or virulence genes, allowing sub-genera level identification and functional analysis.
Figure 1. xGen Amplicon Panels have a single tube workflow that completes in 2 hours. Creating an NGS library starts with multiplex PCR. Your panel is combined with the DNA sample to amplify the targets of
interest. The samples are then amplified with indexing primers to create a functional dual indexed library. As an optional step, the xGen Normalase reagent can be used after pooling multiple libraries to ensure each is equally represented in the final
sample for the flowcell.
Product data
| Features | xGen 16S v2 and ITS1 Amplicon Panels |
|---|---|
| Panel information | 23 primers (16S v2); average 425 bp amplicon size 15 primers (ITS1); amplicon size 145–695 bp |
| Input material | 10 pg for microbial isolates; 1–50 ng for metagenomic samples |
| Time | 2 hours cDNA-to-library or 3 hours cDNA-to-normalized-library-pool |
| Components available |
Note: Kits do not include RT module or magnetic beads |
| Multiplexing capability | Up to 96 combinatorial dual index (CDI) or 1536 unique dual index (UDI) |
| Recommended depth | 16S v2: 100K reads per library ITS1: 25K reads per library |
Coverage of all variable regions of the 16S rRNA gene and ITS1 region
The 16S v2 and xGen ITS1 Amplicon Panels facilitate NGS analysis of complex microbial communities (e.g., bacteria, archaea, fungi) using primer pools that target the 16S rRNA gene (variable regions 1–9) and ITS1 region (Figure 2). In addition, these panels can be customized with additional targets, including antibiotic resistance or virulence genes, allowing sub-genera level identification and functional analysis.
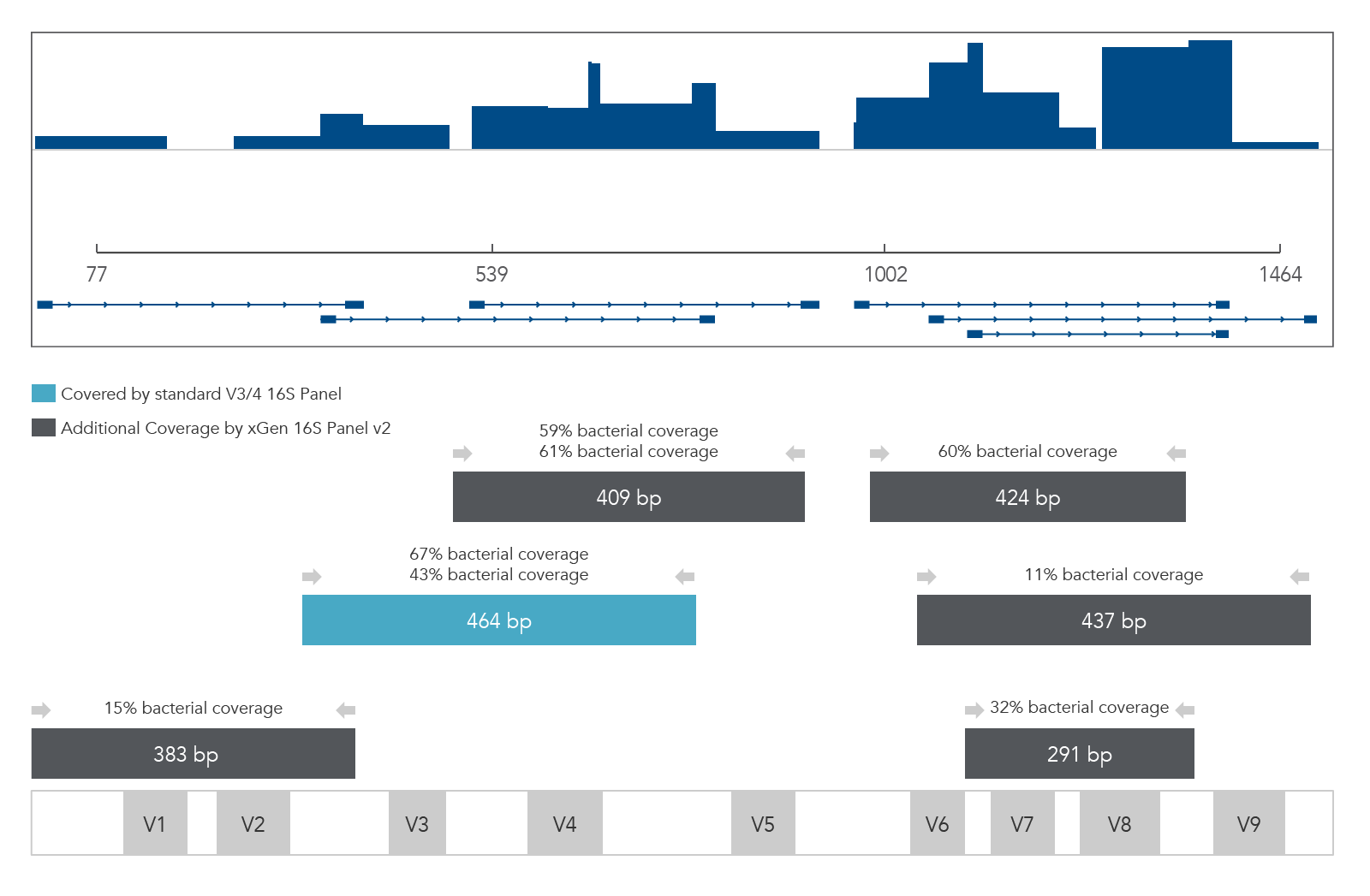
xGen 16S Amplicon Panel v2 (primers)
Figure 2. xGen 16S Amplicon Panel v2 (primers). Sequencing read coverage for an E. coli DNA sample (n = 1) observed in Integrative Genomics Viewer (IGV) Sashimi plot and illustration of multiplexed primer coverage of all nine variable regions of 16S rRNA compared to a standard V3/V4 16S sequencing read coverage.
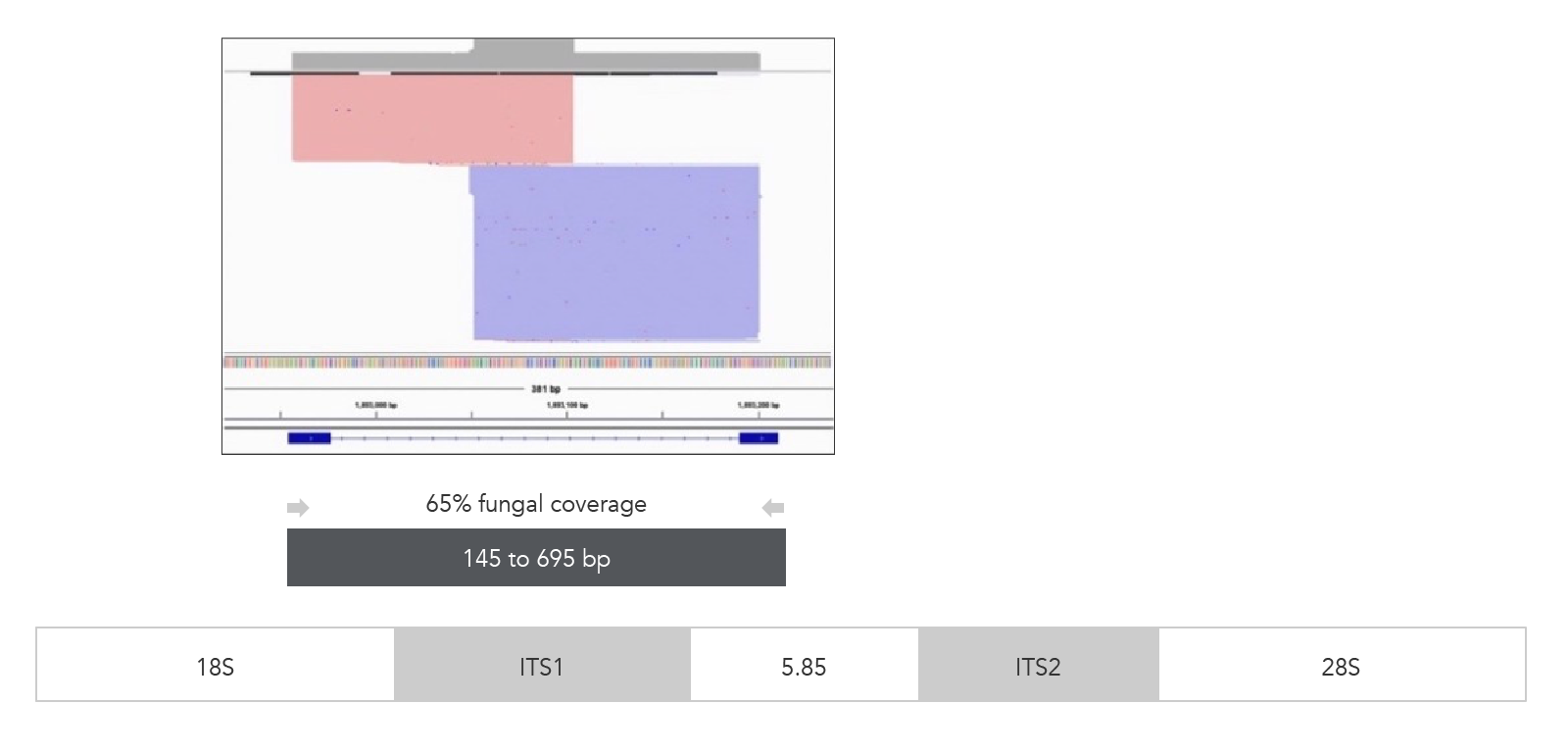
xGen ITS1 Amplicon Panel (primers)
Figure 3. xGen ITS1 Amplicon Panel (primers). Sequencing read coverage observed in the IGV Sashimi plot and illustration of multiplexed primer coverage of ITS1 region for Candida albicans (n = 1). Reads originating from the forward primer are shown in red; reads from the reverse primer are in blue.
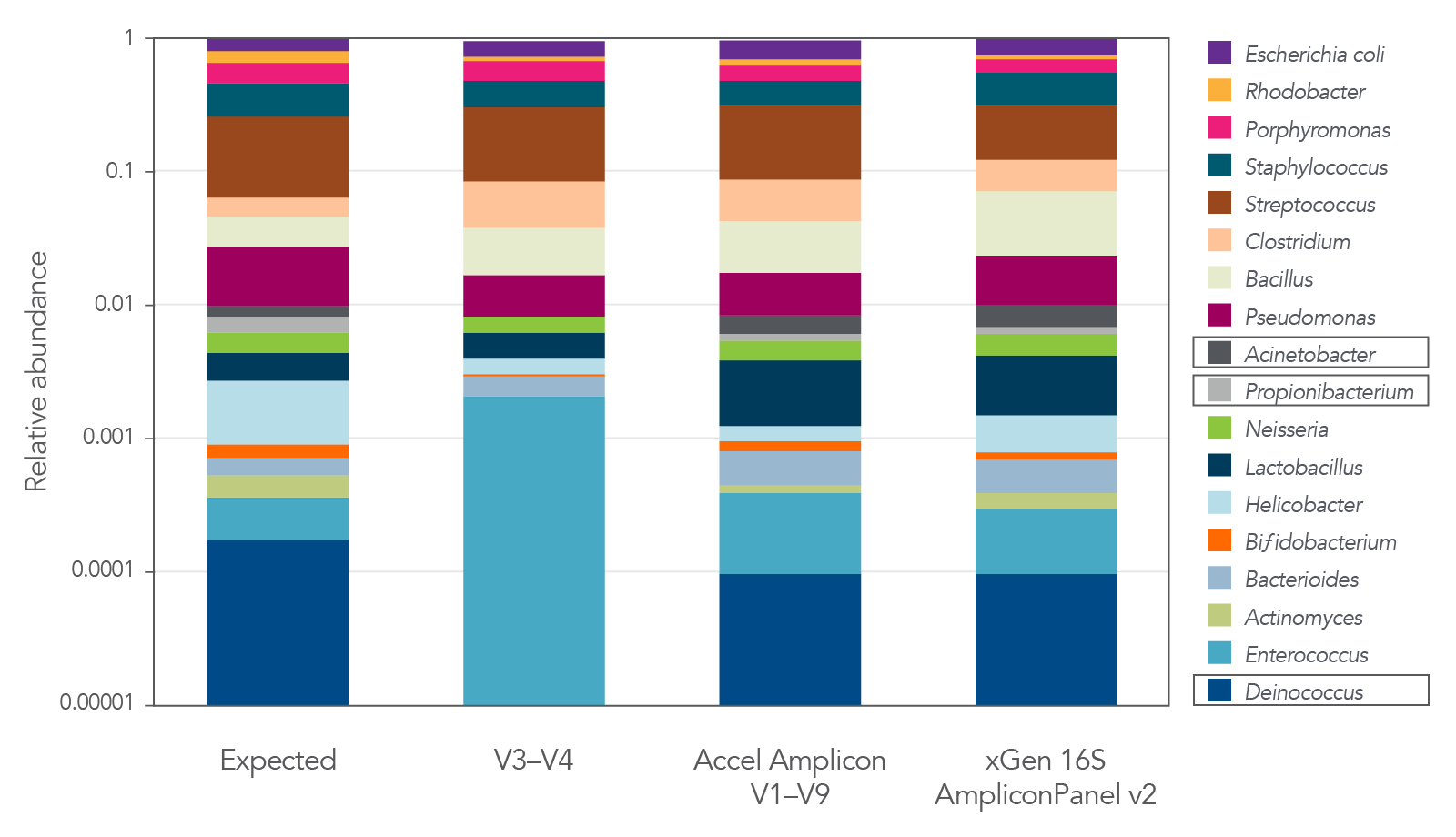
The xGen 16S Amplicon Panel v2 and the xGen ITS1 Amplicon Panel provide representation of diverse microbial communities
Figure 4. The xGen 16S Amplicon Panel v2 provides superior representation of diverse microbial communities. The 16S v2 panel covering V1–V9 regions of 16S rRNA provides accurate representation of each genus in a commercially available standard (MSA-1003) compared to libraries interrogating the V3–V4 region alone. The legacy Accel-Amplicon 16S+ITS panel and the new xGen 16S Amplicon Panel v2 show similar relative abundance results. Strains were present at levels from 0.02% to 18% in MSA-1003. Boxed organisms were not detected with sole use of the V3-V4 region. Libraries were sequenced on a MiSeq® (Illumina) instrument without the use of the PhiX reagent. As shown in the examples in this figure, by not losing reads to PhiX or having to sequence deeper due to the use of phased primers, an increased number of samples can be fit on a sequencing run, thereby increasing sequencing efficiency, while still achieving quality data.
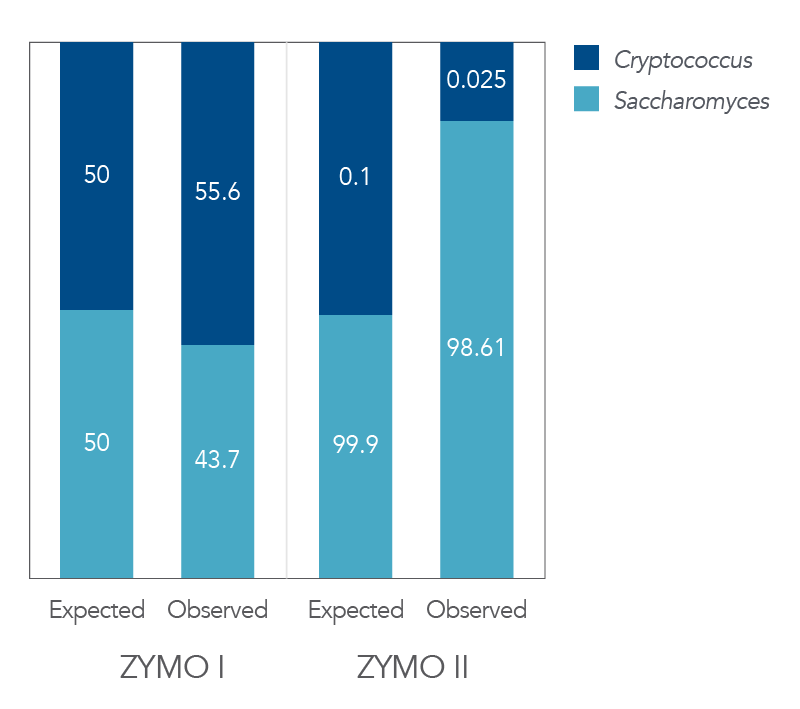
Figure 5. Expected vs. observed Zymo I/II data from the panel covering the ITS1 region. The xGen ITS1 Amplicon Panel provides accurate representation of each genus in two commercially available standards [Zymo I—ZymoBIOMICS™ Microbial Community standard and Zymo II—ZymoBIOMICS Microbial Community standard II (Log distribution)]. Fungal strains were present at levels from 0.1% to 99.9% in Zymo II. The xGen ITS1 Amplicon Panel resulted in expected representation of the fungal species in a bacterial background.
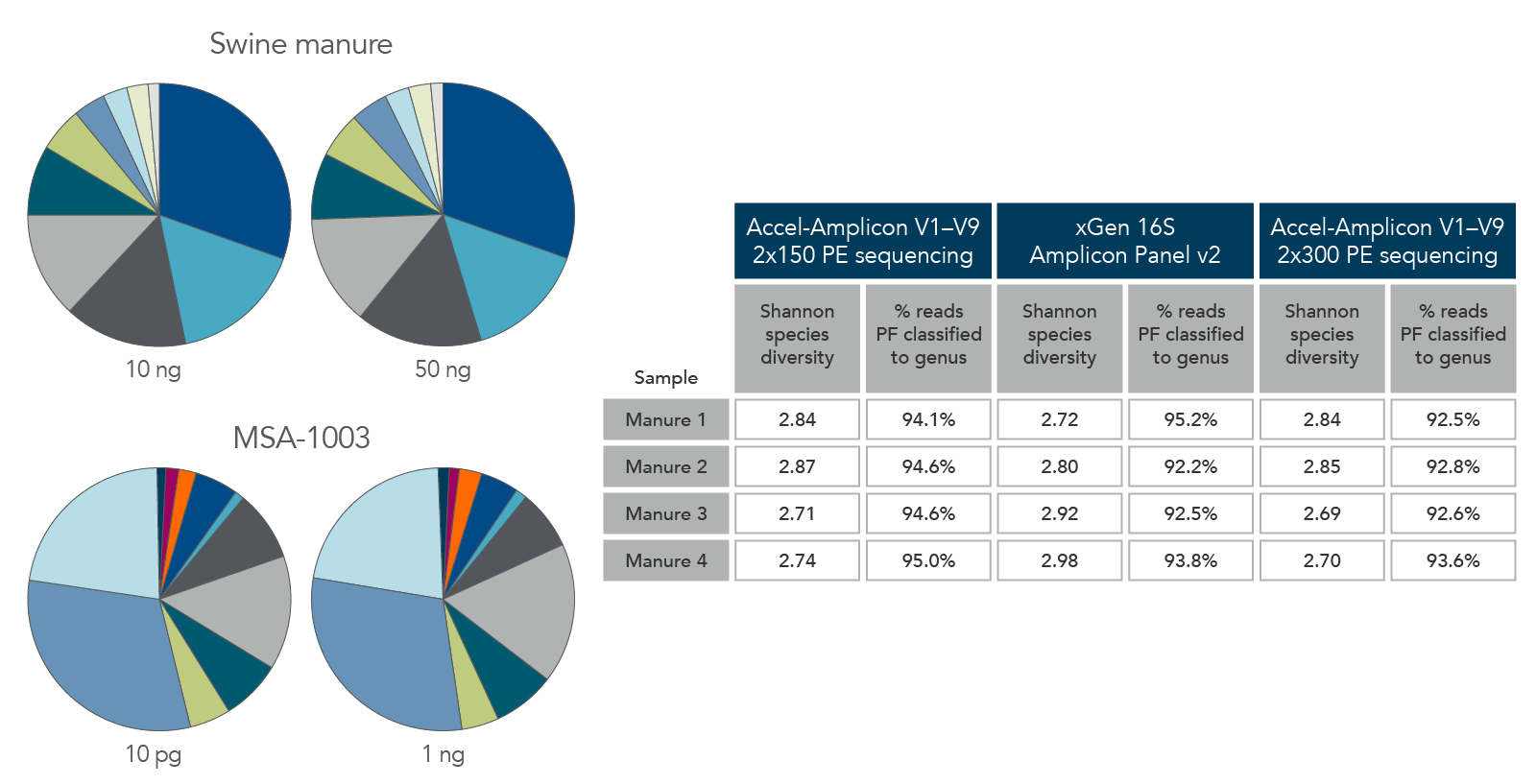
Consistent function with varying biomass, sample types, and read length
Figure 6. Consistent function with varying biomass, sample type, and read length. Using the same protocol and cycling conditions, input quantities of 10 ng to 50 ng of swine manure (top left) identified the same quantities and types of genera. In addition, the 10 pg sample of MSA-1003 (bottom left) had the same relative abundance and type of genera as the 1 ng sample. The legacy Accel-Amplicon 16S+ITS panel and the new xGen 16S Amplicon Panel v2 products gave similar results when comparing the 2 x 150 and 2 x 300 PE sequencing read percentages; a comparable number of genera were identified from swine manure samples (right table).