Advantages of multiplexed sequencing
The capacity of next generation sequencing (NGS) platforms has increased at an astonishing rate. As a result, libraries are commonly pooled together and sequenced simultaneously via a process known as multiplexing. To distinguish individual libraries throughout this process, sample-specific sequences, called sample indexes or sample barcodes, are added to each fragment during library preparation. Libraries can then be pooled and sequenced simultaneously.
Next, the barcode information is used to computationally assign the sequence reads back to the individual libraries. Multiplexing reduces the cost of sequencing substantially and facilitates experimental scalability by amortizing the capture reaction cost across samples being pooled together.
Despite these advantages, multiplexed NGS poses challenges to end users. Sequencing experiments require multiple steps from sample preparation to final data acquisition, and each of these steps can impact final data quality. Here, 2 key metrics are discussed that can act as important indicators of successful multiplexed target enrichment: duplication rate and uniformity. Here are research-based recommendations for successful execution of multiplexed NGS experiments.
Minimizing PCR duplicates
The duplication rate is the fraction of mapped reads where any 2 reads share the same 5′ and 3′ coordinates. Duplicates mostly arise from the PCR step during library construction. Duplicates may also result in artifacts on the sequencing instrument where the same template binds to multiple clusters on a flow cell. This results in the same template being amplified independently multiple times across the clusters. Both types of duplications are an important source of error because the resulting reads may contain mutations introduced during the PCR step. These mutations can generate errors when measuring allele frequency representation by increasing the proportion of the duplicated allele compared to other alleles [1].
Many analysis pipelines remove PCR duplicate reads before downstream analysis to mitigate these undesired consequences and minimize potential variant calling biases. Picard (MarkDuplicates; [2]) and SAMTools (rmdup; [3]) are 2 main software programs used for this purpose. The removal of duplicates, however, leads to fewer sequences per sample. Having fewer reads and low-quality samples in this manner increases the cost of sequencing due to duplicates. Thus, minimizing duplicates during NGS library preps is critically important for PCR-amplified sequencing libraries [4].
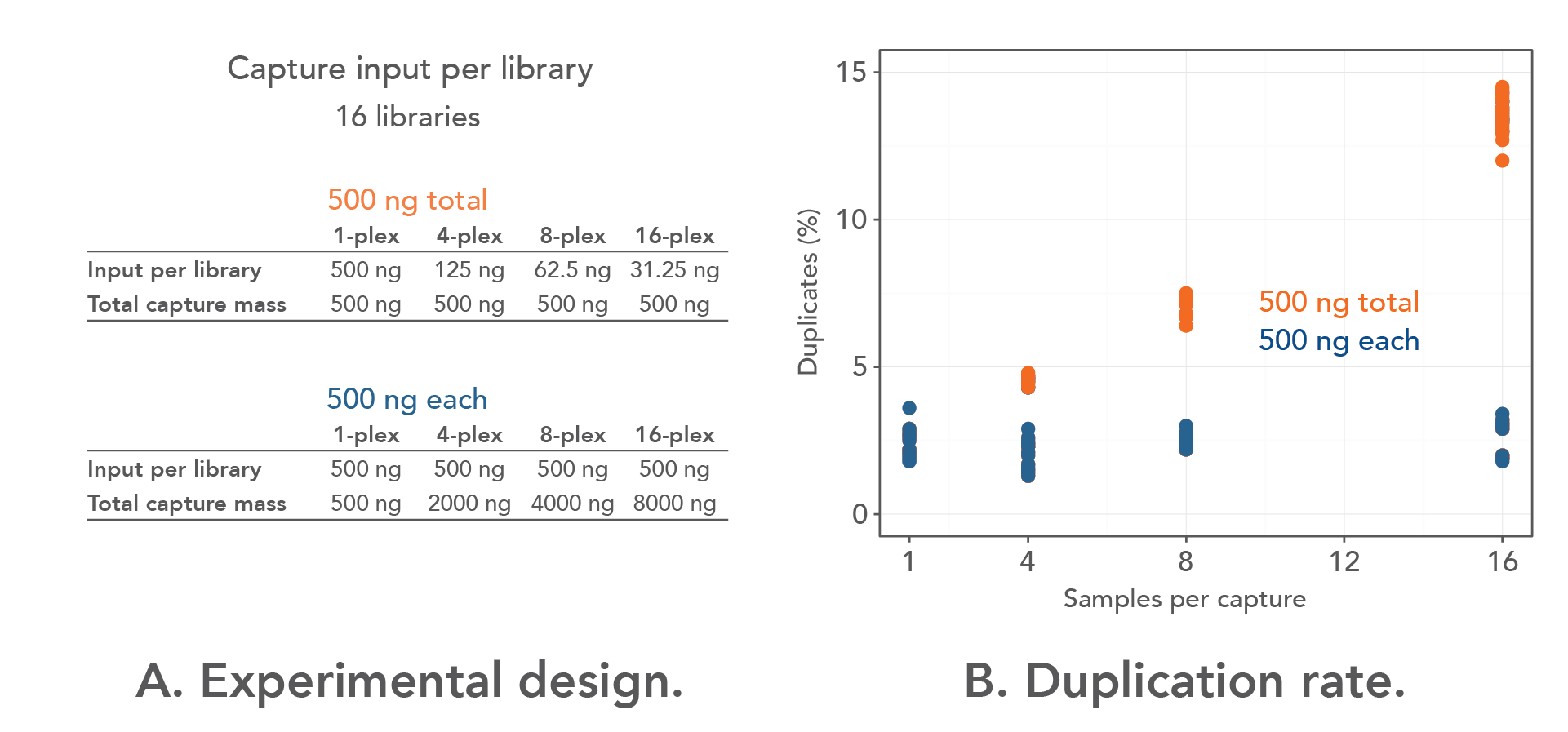
The amount of starting material that is pooled during hybridization capture plays an important role in determining the rate of duplication in multiplexed NGS experiments [5]. To determine the amount of barcoded library needed to minimize duplicates in multiplexed capture, 16 libraries were prepared from Coriell DNA (NA12878) using custom, dual-indexed adapters with 8 nt indexes (IDT) and a T/A ligation based library prep kit. One-, 4-, 8-, and 16-plex pools were then captured with either 500 ng of total input or 500 ng of each library (Figure 1A) using the IDT xGen™ AML Cancer Hyb Panel (1.19 Mbp) . For example, the 16-plex captures contained either 31.25 ng of each library, totaling 500 ng per capture, or 500 ng of each library, totaling 8 µg per capture. Importantly, no other modifications were made to the experiments, and the same amount of hybridization capture panel, blockers, and DNA for multiplexed captures was used.
As shown in Figure 1B, the duplication rate was consistent when the libraries were captured individually (2.4%). However, there was an increase in the duplication rate in the "500 ng total input" groups (orange circles) when the libraries were captured in 4-plex (4.5%) instead of 1-plex (2.0%). The rate of duplication increased substantially in the same groups when the libraries were captured in 8-plex (7.1% vs. 2.4%). The biggest increase in duplication rate was observed when capture was performed in 16-plex with the 500 ng total groups (13.5% vs. 2.5%). Importantly, through the experiments, the duplication rate remained almost constant in the "500 ng each library" groups (blue circles), whether they were captured individually or in multiplex (4-plex, 8-plex, or 16-plex).
Based on these data, 500 ng of each barcoded library is recommended to be used in multiplexing experiments to reduce PCR duplicates.
High coverage uniformity with multiplexed captures
Sequencing coverage or coverage depth represents the number of times sequencing reads “map to” or “cover” a genomic target region. Coverage level impacts the ability to find sequencing variants. A higher sequencing coverage increases the ability to confidently identify novel variants. The coverage level required for an experiment depends on factors such as application type (SNPs, mutations, genomic rearrangements) and expression level of target genes (low or high expression genes for RNA-seq).
In many applications, successful targeted sequencing also requires uniform coverage across the regions of interest within the genome. Generally, each target site is desired to be covered at the same level, which would keep the required number of sequencing reads for every target site at the minimum. However, sequencing reads are often not distributed evenly over the target areas, meaning that extra reads and thus extra sequencing is required to “rescue” the poorly covered regions. Thus, obtaining high uniformity of coverage is essential for a cost-effective multiplex sequencing experiment aiming to identify novel variants.
To determine whether using 500 ng of each library in multiplexing experiments provides uniform coverage, the per-base target coverage was examined in the previously described experiment. As seen in Figure 2, target coverage was highly uniform, regardless of the number of samples multiplexed. Base coverage was 98.2% for 20X for all 4 experimental groups. An average of 94.8% of the bases were covered at least 100X. Coverage was nearly 200X for 61.8% and 300X for 23.6% of bases.
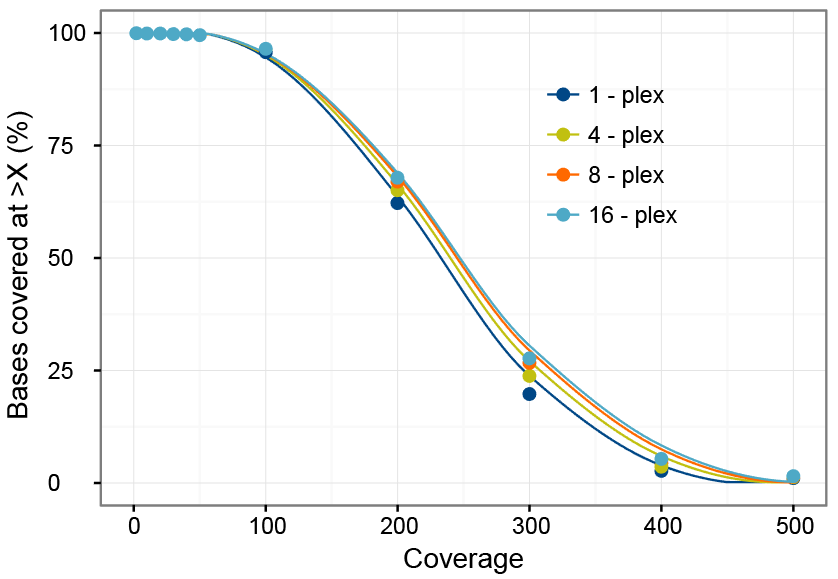
These results suggest that pooling of 500 ng per library, captured using the xGen AML Cancer Hybridization Panel provides high coverage uniformity and high target coverage enabling variant calling with minimal sequencing in multiplexed NGS experiments.
Other considerations
It is noteworthy to mention that several other factors, including sample quality, PCR conditions, panel size, and the number of samples multiplexed should be studied carefully in your experiments.
Our scientific application specialists are available to answer further questions or provide guidance on sample multiplexing for your NGS experiments. Our scientific application specialists are available to answer further questions or provide guidance on sample multiplexing for your NGS experiments, please contact them here.

