Guide RNA design: Be on target!

Here are general considerations for guide RNA design:
On- and off-target activity
The goal when designing a guide RNA is achieving the highest possible on-target activity of your CRISPR gene editing complex, while minimizing off-target activity. Off-target activity causes unwanted phenotypes, potentially including cell death.
Cas9 guide RNA design considerations
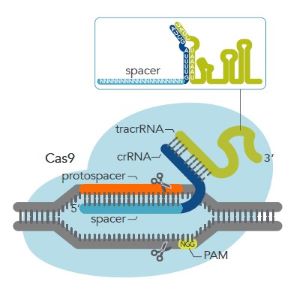
For Cas9 guide RNA designs, the target sequence must be next to a PAM sequence, NGG, where N is any base. It is important that you do not include the PAM sequence in the actual guide RNA design. The guide RNA recognizes and binds to 20 nucleotides on the DNA strand opposite from the NGG PAM site. The “N” of the NGG is immediately adjacent to the most 3’ base of the non-targeted strand side of the protospacer (see Figure 1 below).
Figure 1: PAM site and Cas9 gRNA. The PAM site is on the non-targeted strand of DNA. The guide RNA binds to the targeted strand. The guide RNA in this figure is a 2-part guide RNA consisting of crRNA (light blue and dark blue) and tracrRNA (green).
Cas9 guide RNA length considerations
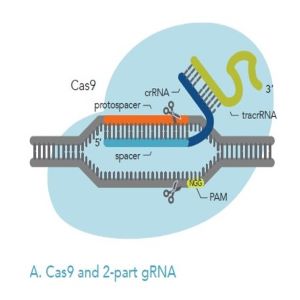
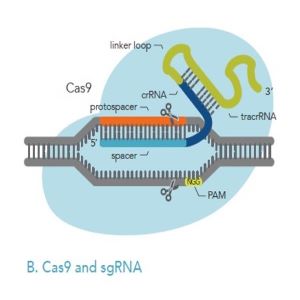
If you decide to use Cas9 and chemically synthesized guide RNA, you have the choice of a 2-part or single guide RNA (Figures 2A, 2B below). Both guide RNA formats are commonly delivered to cells as Cas9 RNP.
Figures 2A, 2B: Comparison of 2-part gRNA (A) and sgRNA (B). Both kinds of gRNA can be added to Cas9 to form RNP, which can then be delivered to cells.
When you are using 2-part guide RNA, there are crRNA and tracrRNA components:
crRNA: The crRNA is composed of a target-specific spacer region (light blue) and another domain (dark blue) that hybridizes to the tracrRNA (green). The target-specific spacer sequence is 20 nucleotides long; if it is shorter than this, the on-target activity will be negatively impacted. IDT researchers have found that the optimal total length of the crRNA (the target-specific spacer region plus the domain that hybridizes to tracrRNA) is 36 nucleotides.
tracrRNA: Part of the tracrRNA molecule hybridizes (base pairs) to the crRNA, and another part of it binds to Cas9. IDT scientists have found that the optimal, total length for tracrRNA is 67 nucleotides. This is shorter than the tracrRNA found in nature, but IDT scientists have found that when using 2-part guide RNA, shortening the tracrRNA to 67 nucleotides increases on-target activity.
When using sgRNA, the crRNA and tracrRNA are all part of one longer RNA molecule joined by a hairpin-like loop (linker-loop, Figure 1). IDT scientists have found that the optimal length for sgRNA for genome-editing purposes is 100 nucleotides.
If you need help choosing which type of guide RNA is best for your CRISPR Cas9 experiment, this DECODED article provides more discussion and guidance about which gRNA format to use (quick tip: it usually does not make a difference).
CAUTION: Cas9 guide RNA design - The CRISPR-Cas9 complex can tolerate several mismatches between the guide sequence and the target sequence, meaning it is fairly easy to end up with undesirable off-target effects. Thus, we recommend using high-quality guide RNA design software.
Cas12a guide RNA design considerations
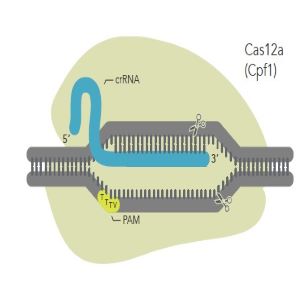
For Cas12a guide RNA designs, just like for Cas9 guide RNA, the target sequence must be next to a PAM sequence. But for Cas12a, the PAM sequence is TTTV, where V can represent A, C, or G. In addition, if you are using Alt-R Cas12a Ultra, a TTTT PAM sequence may also work but may not be as potent.
The “V” of the TTTV is immediately adjacent to the most 5’ base of the non-targeted strand side of the protospacer element (Figure 3 below). As for Cas9 guide RNA designs, it is important that you do not include the PAM sequence in the guide RNA design when ordering.
The guide RNA for Cas12a is relatively short at only approximately 40-44 bases long. The part that base pairs to the protospacer is 20-24 bases in length, and there is also a constant 20-base section that binds to Cas12a (Figure 3 below). Our scientists have found the optimal total length to be 41 bases (21 target-specific bases that will base pair with the protospacer, and 20 constant bases for binding the Cas12a enzyme).
Figure 3: Cas12a and its crRNA. The crRNA for Cas12a is relatively short and does not require tracrRNA.
NOTE: gRNA nomenclature - Guide RNA for Cas12a is often referred to as crRNA, even though there is no tracrRNA.