Digital PCR
Overview
Digital PCR (dPCR) provides the absolute amount of a target DNA. Properly designed assays can be used to investigate rare alleles, rare microRNAs, copy number variations, and low abundance transcripts. Digital PCR applications span many scientific fields, from liquid biopsy analysis for oncology research to viral load assessment for infectious disease research. Although the underlying reaction chemistry shares many similarities with qPCR, digital PCR offers distinct advantages and complexities.
What is digital PCR?
Digital PCR (dPCR) quantifies the absolute number of copies of a target genomic or complementary DNA (cDNA) sequence from a sample and can be used for a variety of applications [1–3]. For genotyping applications, dPCR can capture rare allele or copy number variation, be used for liquid biopsy analysis, identify homology directed repair (HDR) or non-homologous end joining (NHEJ) induced by CRISPR-based genome editing, and assess DNA quality for next generation sequencing (NGS). dPCR can also assess gene expression from single cells, determine expression levels of microRNAs, and measure viral load. Although the reagents are very similar to those used for traditional quantitative PCR (qPCR), the method of quantification is different. Some key similarities and differences between dPCR and qPCR are outlined in the following breakdown of dPCR steps.
Digital PCR steps and applications
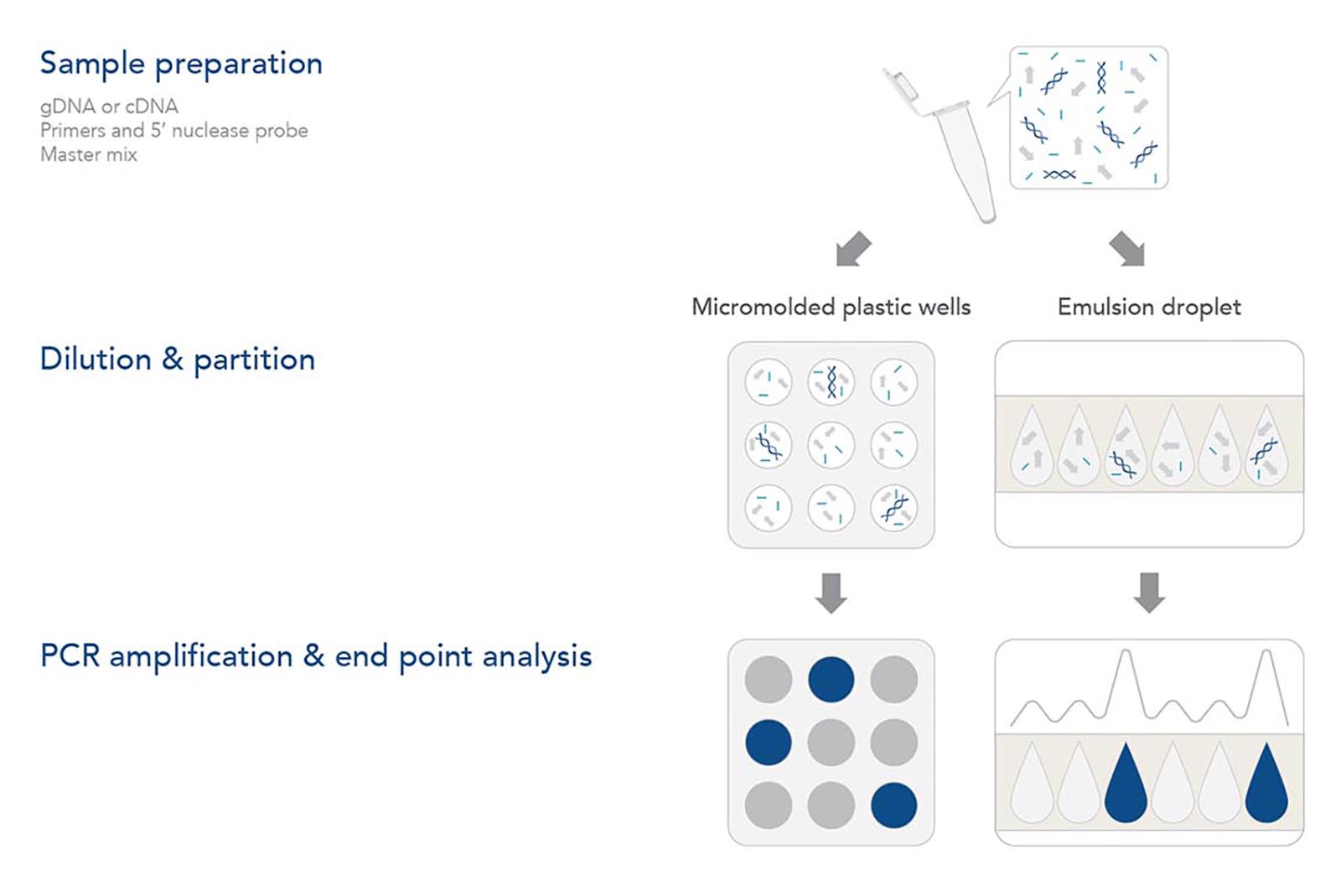
Step 1. Sample preparation
As with any genomic analysis, a sample of nucleic acid must be obtained. For genotyping applications, a sample of the genomic DNA (gDNA) needs to be purified from the tissue or sample of interest. As with qPCR, dPCR amplifies the target DNA using a thermostable DNA polymerase and is unable to amplify RNA directly. Thus, gene expression analysis requires mRNA from the sample of interest to be isolated and converted into cDNA with reverse transcriptase.
After purified gDNA or cDNA is obtained, the sample is mixed with forward and reverse primers and a hydrolysis (5' nuclease) probe. Each of these sequences are designed to have sequence complementarity to the target gene/DNA of interest. As with qPCR, the primers anneal to the outer regions of the target sequence (typically 60–150 bp in length). The 5′ nuclease probe—containing a 5' fluorophore and a 3' quencher—anneals between the forward and reverse primers to help identify the PCR target in the reaction. Hydrolysis probes include a 5′ fluorophore and 3′ quencher. As in a traditional qPCR assay, the primer and probe design are key to obtaining reliable results. More information on the design of these components can be obtained by reviewing the Real-time PCR guide: Part 1—assay design. IDT offers a variety of different products to support your dPCR experiments, including PrimeTime™ qPCR Probes, PrimeTime qPCR Probe Assays, and Affinity Plus™ qPCR Probes with locked nucleic acids.
Digital PCR can also be performed with intercalating double-stranded DNA dyes, such as SYBR® Green (Life Technologies) or EvaGreen® (Biotium), rather than 5′ nuclease probes [4]. The design of primers for these assays is critical since the amplification of primer-dimers cannot be distinguished from target amplification. Using well-established protocols and performing melt-curve analysis on the products can help delineate target amplification as opposed to other PCR products. For more information see the IDT webinar, Understanding melt curves for improved SYBR Green assay analysis and troubleshooting.
Step 2. Dilution & partition
Although many reagents used for digital PCR are identical to qPCR assays, several aspects of the reaction and data generation stage differ. dPCR experiments first partition the reaction into individual nanoliter reactions with individual partitions containing one (or very few) target DNA molecules while others contain none. Two distinct methods are used to partition the reaction. In some dPCR platforms, the reaction is divided into separate chambers, such as individual micromolded plastic wells. Other dPCR platforms create nanoliter sized reaction droplets as an oil/water emulsion. Rather than analyzing a single amplification reaction mixture in real time like qPCR, dPCR analyzes ~20,000 individual qPCR reactions.
Step 3. PCR amplification & end point analysis
After partitioning, the plate or water droplets in the water/oil emulsion are amplified using regular PCR cycling parameters. As DNA polymerase extends from the forward primer during the elongation/extension part of the cycle, its exonuclease activity degrades the probe, releasing the 5′ fluorophore from the 3′ quencher and enabling fluorescence. If the assay includes intercalating dyes rather than a 5' nuclease probe, then the fluorescence increases as double-stranded PCR amplicons accumulate.
After the PCR cycle is complete, any partition that included at least 1 template of target DNA sequence will fluoresce. Any droplet or well that was devoid of the target DNA will not fluoresce. The total number of droplets or wells with fluorescence represents the total number of target molecules in the sample. Statistically, there are opportunities for some partitions to have more than 1 target DNA that are accounted for by applying the Poisson distribution function when determining the total number of target molecules in the sample [5].
Multiplex Digital PCR
The individual partitions are also amenable to multiplexing, a process where multiple target DNAs or cDNAs are analyzed simultaneously. Just like a multiplex qPCR reaction, 2–5 sets of primers and probes with distinct fluorophores are mixed with the sample. Since the template or target DNAs are partitioned into individual reactions, one set of primer and probes will amplify the individual target; therefore, the primers and probes will not compete for reagents during amplification as in a typical multiplex qPCR assay. During analysis, the number of partitions for each of the fluorophores represents the number of individual targets for that particular primer and probe set. Counting the partitions for each primer and probe set in the multiplex assay can establish an absolute ratio for the different targets. Multiplexing dPCR reactions can also facilitate the quantification of rare alleles for a gene and determine copy number variations.
Table 1. Digital PCR instrument compatibility and reporter dyes.
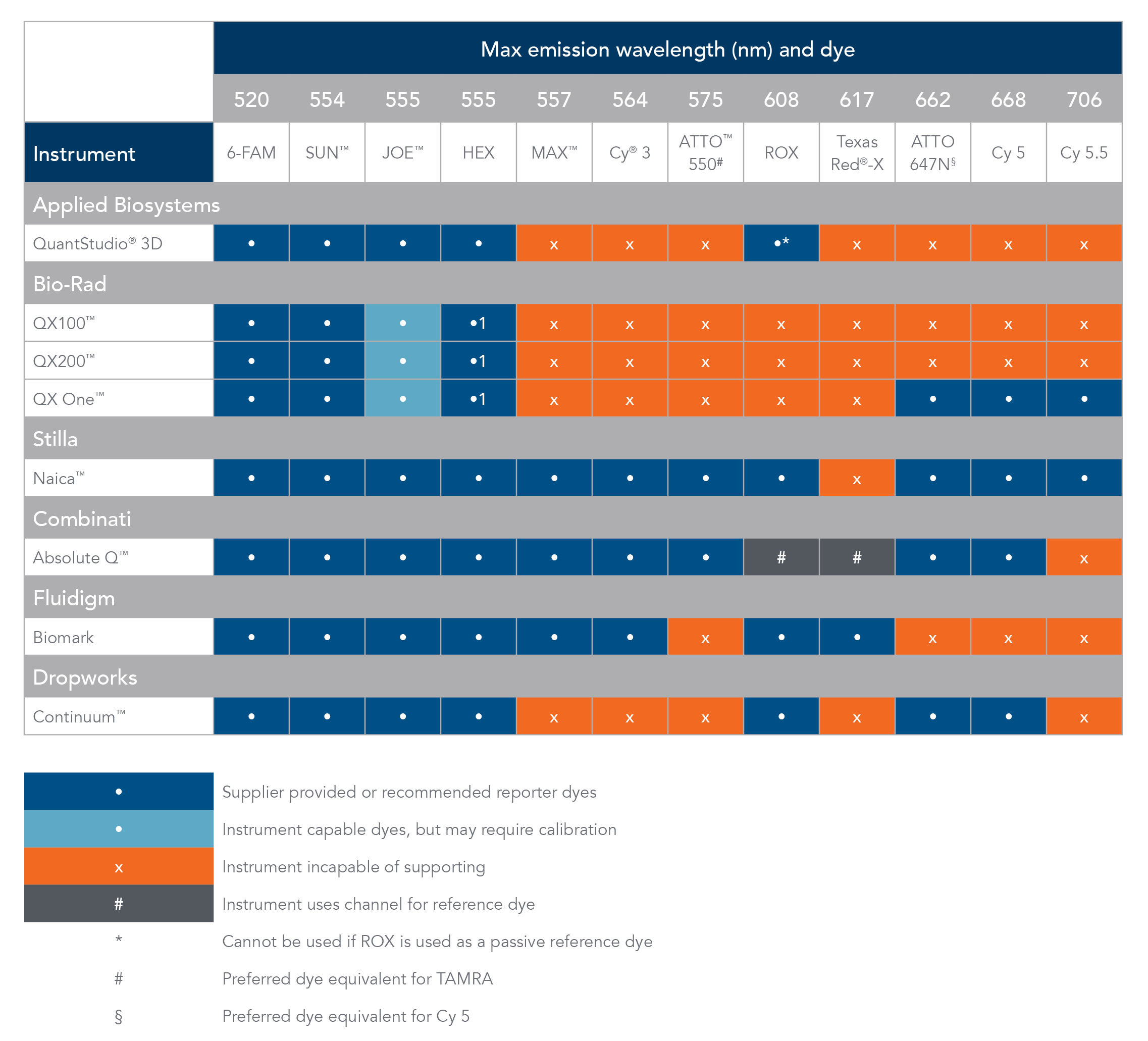
Cy is a registered trademark of Cytiva. Texas Red and VIC are registered trademarks of Life Technologies. ATTO is a trademark of ATTO-TEC GmbH. QuantStudio 3D and JOE fluorophore are trademarks of Thermo Fisher Scientific. QX100, QX200, and QX One are trademarks of Bio-Rad. Naica is a trademark of Stilla Technolgies. Absolute Q is a trademark of Combinati. Continuum is a trademark of Dropworks.
Probes and assays for digital PCR
Cost-effective and conveniently-packaged reagents for genotyping and gene expression applications using dPCR.
Get started with digital PCR
Ready to use digital PCR in your research program? IDT offers a wide variety of predesigned and customizable qPCR products well-suited for digital PCR applications. Design an efficient digital PCR workflow for genotyping or gene expression analysis with our flexible ordering options, such convenient size offerings and customizable probes. Check out our PrimeTime™ and Affinity Plus™ product lines or contact us today to learn more about getting started with digital PCR.
Explore digital PCR resources
References
1. Hindson BJ, Ness KD, Masquelier DA, et al. High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number. Anal Chem. 2011;83(22):8604-8610.
2. Morley AA. Digital PCR: A brief history. Biomol Detect Quantif. 2014;1(1):1-2.
3. Altgilbers S, Dierks C, Klein S, Weigend S, Kues WA. Quantitative analysis of CRISPR/Cas9-mediated provirus deletion in blue egg layer chicken PGCs by digital PCR. Sci Rep. 2022;12(1):15587.
4. Tai AC, Parfenov M, Gorham JM. Droplet Digital PCR with EvaGreen Assay: Confirmational Analysis of Structural Variants. Curr Protoc Hum Genet. 2018;97(1):e58.
5. Basu AS. Digital Assays Part I: Partitioning Statistics and Digital PCR. SLAS Technol. 2017;22(4):369-386.
*RUO—For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend for these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations.