Whole genome sequencing
Succeeding in your whole genome sequencing (WGS) research is critical to improving the lives of all living things—today and well into the future. Accelerate your research with our xGen whole genome sequencing solutions.
xGen™ NGS—made to reach.
Overview
- Recommended products—curated for whole genome sequencing
- Short workflow—create a normalized library pool in under 4 hours
- Low input, high complexity—up to 3-fold fewer duplicates from 1 ng of DNA when using the xGen DNA Library Prep Kit EZ
- Enzymatic fragmentation—no need for specialized equipment or consumables for DNA fragmentation
- xGen Normalase™ technology—a proprietary enzymatic method of library normalization for multiplexed sequencing
- Automation friendly—works on a variety of platforms
What is whole genome sequencing?
Whole genome sequencing provides comprehensive coverage over the entire mappable genome to gain information about an organism or metagenomic DNA samples. In comparison to targeted sequencing that is often limited to a subset of genes of interest, WGS includes coverage of all coding and non-coding regions, regulatory sequences, and inter- and intra-genic portions of the genome. Then, the sequencing data are aligned to a reference genome for variant analysis, assembled into contigs for de novo genome assemblies, or used for microbial classification (in the case of metagenomic samples).
For low-throughput labs, the 16-reaction product solution for whole genome sequencing is available that includes library prep and adapters. This solution offers a highly streamlined workflow that consists of only three steps:
- Enzymatic preparation—DNA fragmentation, end-repair, and dA-tailing of dsDNA
- Adapter ligation
- PCR amplification
For library prep, the xGen DNA Library Prep Kit EZ uses an enzymatic fragmentation strategy to shear high-quality DNA samples. The kit employs TA ligation-based library prep and includes an enzyme mix for fragmentation, end-repair and dA-tailing, a ligase to attach the xGen Stubby Adapter, as well as a high-fidelity polymerase for indexing PCR. This solution offers premixed xGen UDI Primer Pairs (not included with the xGen DNA Library Prep Kit EZ) for indexing by PCR, which generates an NGS library ready for Illumina® sequencing.
For high-throughput labs, the 96-reaction solution can be used along with the xGen Normalase Module (sold separately), a proprietary enzymatic library normalization kit for multiplexed sequencing. The method doesn't require individual library quantification and utilizes equal volume pooling instead of requiring a different volume for each library when making an equimolar library pool for multiplexed sequencing. The Normalase enzymatic normalization also results in more uniform read depth within a library pool, with a coefficient of variation (CV) <10% when compared to methods that require individual library quantification
Extraction
LIBRARY PREP
Library prep kits
-
xGen DNA Library Prep Kit EZ (16 or 96 rxn)
for high-quality DNA samples
Indexing primers
- UDI primers (16 rxn)
-
UDI Normalase primers (96 rxn)
Normalization
xGen Normalase Module (96 rxn)
Sequencing & analysis
Method data
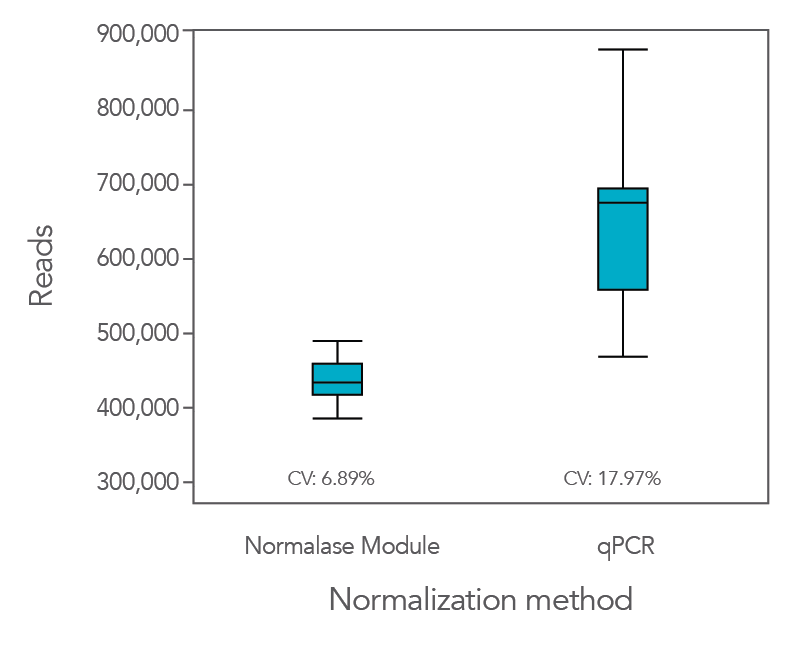
Normalase leads to the formation of balanced library pools without quantification resulting in uniform sequencing data
Normalization using the proprietary Normalase workflow in comparison to the traditional qPCR quantification method provides more uniform results, maximizing the value of the sequencing data as it provides a tighter grouping of reads between samples.
In an experiment where the xGen DNA Library Prep Kit EZ was used in conjunction with Coriell NA12878 genomic DNA (gDNA), xGen Stubby Adapter, xGen UDI primers, and Normalase, the CV was lower (6.9%) compared to when libraries were quantified by qPCR (18%,
Figure 1). This result indicates that the use of Normalase provides a faster, more efficient workflow for multiplex sequencing.
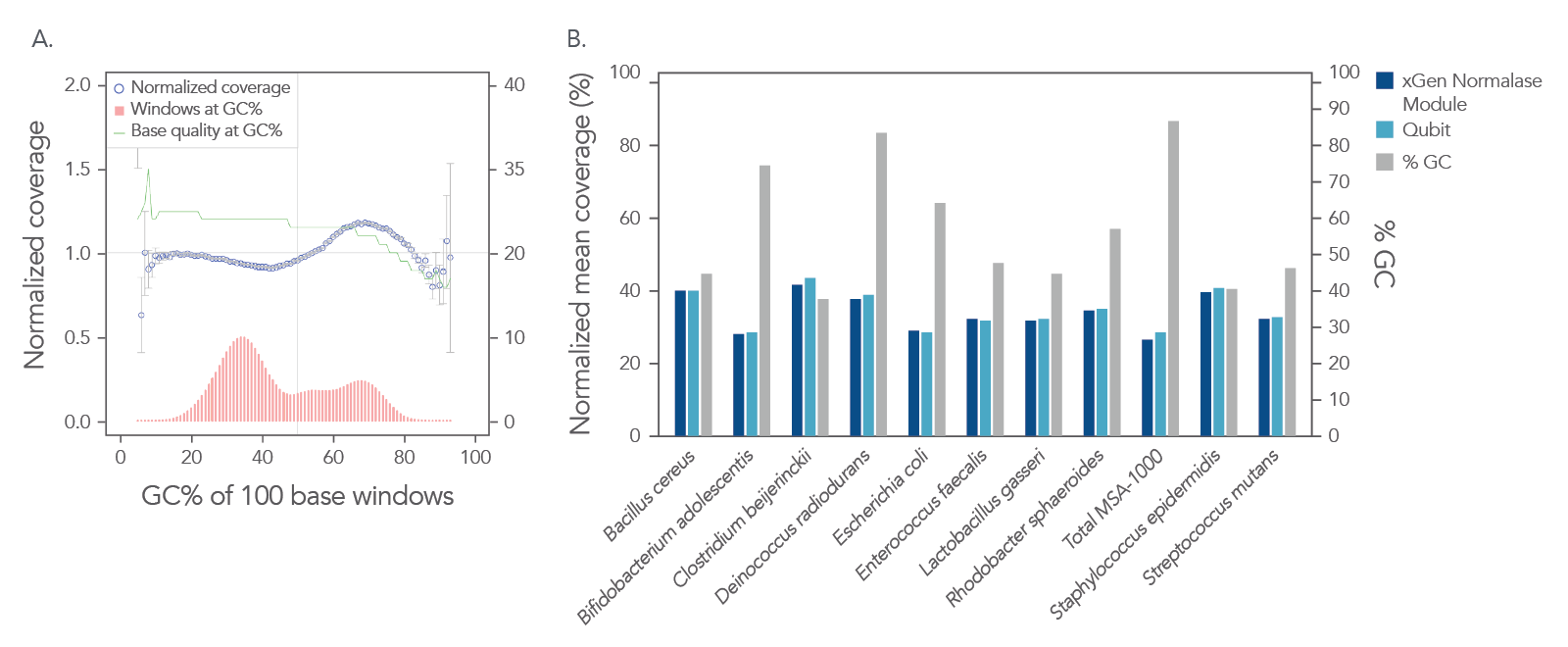
In a shotgun metagenomic sequencing experiment, libraries were constructed using the xGen DNA Library EZ Kit and 1 ng of DNA derived from a mock metagenome DNA standard (ATCC, MSA-1000™) with varying GC content (30–68.8%). The kit provided
comprehensive normalized coverage across distinct GC compositions and coverage uniformity (Figure 2A). The data also show that the libraries pooled using the Normalase workflow maintain a high-quality sequencing coverage across a wide range of
GC targets (Figure 2B).
Figure 1. The Normalase workflow enables streamlined library balancing and pooling process without the need to quantify samples and maximizes sequencing efficiency. xGen DNA Library Prep EZ libraries were generated with stubby adapters from 100 ng of Coriell NA12878 gDNA. Twelve library subsets were either pooled and normalized based on qPCR quantification or pooled and normalized using the xGen Normalase Module, followed by co-sequencing to determine the percentage of reads corresponding to each sample index using MiSeq® V2 Standard 300-cycle kit (Illumina).
Figure 2. Normalized coverage, coverage uniformity (A), and strain specific normalized coverage (B). xGen DNA libraries (n = 4) were made from 1 ng inputs of ATCC MSA-1000™ DNA—an equal mass mix of 10 different bacterial strain genomes with varying GC%. Two libraries were amplified and indexed either with xGen CDI Normalase primers or IDT CDI primers. Normalase conditioned libraries were normalized to 4 nM and pooled using Normalase, while standard libraries were quantified using Qubit™ (Thermo Fisher Scientific) and pooled at 4 nM. Libraries were co-sequenced on a MiniSeq (Illumina) at 2 x 150 bp read length. Reads were subsampled to 5 M per sample. Normalase-treated libraries maintained high-quality genomic coverage across bacterial genomes with varying GC%. The ATCC MSA-1000™ DNA standard contains the following strains: 10% Bacillus cereus (ATCC 10987), 10% Bifidobacterium adolescentis (ATCC 15703), 10% Clostridium beijerinckii (ATCC 35702), 10% Deinococcus radiodurans (ATCC BAA-816), 10% Enterococcus faecalis (ATCC 47077), 10% Escherichia coli (ATCC 700926), 10% Lactobacillus gasseri (ATCC 33323), 10% Rhodobacter sphaeroides (ATCC 17029), 10% Staphylococcus epidermidis (ATCC 12228), 10% Streptococcus mutans (ATCC 700610).